Projecting the graph on a subset of strains.
In this final part of the tutorial we show how to use the marginalize command. This command is used when one is interested in the relationship between a subset of the genomes that make up the pangraph. It can quickly create, starting from the full pangraph, a smaller pangenome graph relative to the selected subset of strains.
Preliminary steps
We will run this tutorial on a different dataset, containing 10 complete chromosomes of Klebsiella Pneumoniae (source: GenBank). These sequences are available in the pangraph repository (example_dataset/klebs.fa.gz) and can be downloaded by running:
wget https://github.com/neherlab/pangraph/raw/master/example_datasets/klebs.fa.gzAs for the previous dataset, we can create the pangraph with the command:
pangraph build --circular klebs.fa.gz > klebs_pangraph.jsonOn 4 cores the command should complete in around 6 mins. After creating the pangraph, we can export it in gfa format for visualization.
pangraph export \
--no-duplications \
--output-directory klebs_export \
--prefix klebs_pangraph \
klebs_pangraph.jsonThis will create the file klebs_export/klebs_pangraph.gfa which can be visualized using Bandage.
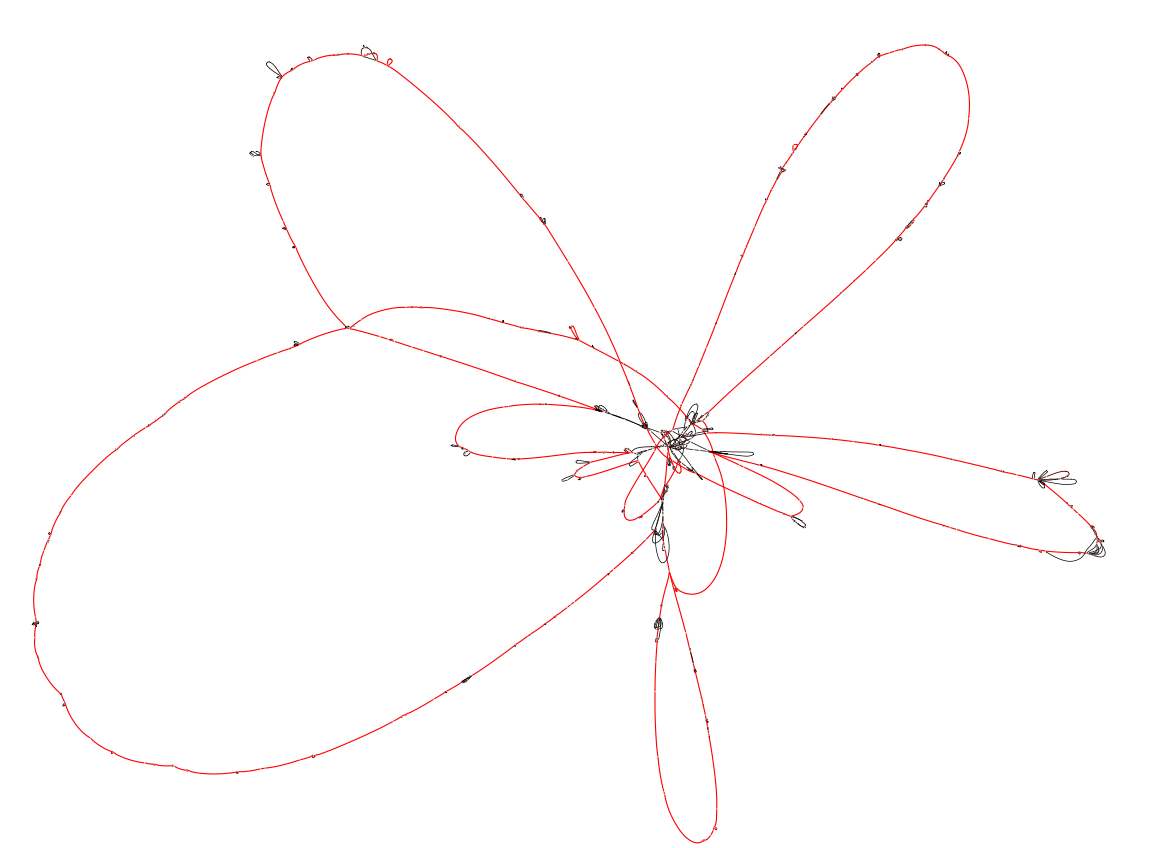
Colors indicate the number of times a block occurs. Blocks that appear in red are core blocks that are found in every chromosome, while black blocks are only present in a few strains. We used the --no-duplication flag in the export command, which excludes duplicated blocks from the exported graph. This simplifies the visualization, which would otherwise be highly "tangled-up" by these duplications.
Marginalize the graph on a subset of strains
The full graph might be difficult to interpret. However if we are interested only in relationships between a subset of chromosomes we can use the command marginalize (see Marginalize) to project the pangraph on this set of strains. This "marginalization" operation will remove transitive edges. If two blocks always come one after the other (with same strandedness) in all of the subset of strains one is interested in, these blocks will be merged in a new larger block. This can greatly simplify the pangenome graph, and highlight differences between a particular subset of strains. And it is also computationally much faster than building a new pangraph directly from the sequences of interest.
For this example we will consider the pair of strains NZ_CP013711 and NC_017540.[1] We can marginalize the pangraph on these two strains by running:
pangraph marginalize \
--strains NZ_CP013711,NC_017540 \
klebs_pangraph.json > klebs_marginal_pangraph.jsonThe file klebs_marginal_pangraph.json will contain the new marginalized pangraph. The strains on which one projects are specified with the flag --strains. They must be passed as a comma separated list of sequence ids, without spaces.
If the command marginalize is executed without specifying particular strains, but providing an output directory with the option --output-path, then it will automatically produce all pairwise marginalizations and save them in the specified directory.
A look at the marginalized pangraph
As done for the main pangraph, we can export the marginalized pangraph in gfa format:
pangraph export \
--no-duplications \
--output-directory klebs_export \
--prefix klebs_marginal_pangraph \
klebs_marginal_pangraph.jsonThis will produce the file klebs_export/klebs_marginal_pangraph.gfa which can be visualized with Bandage.
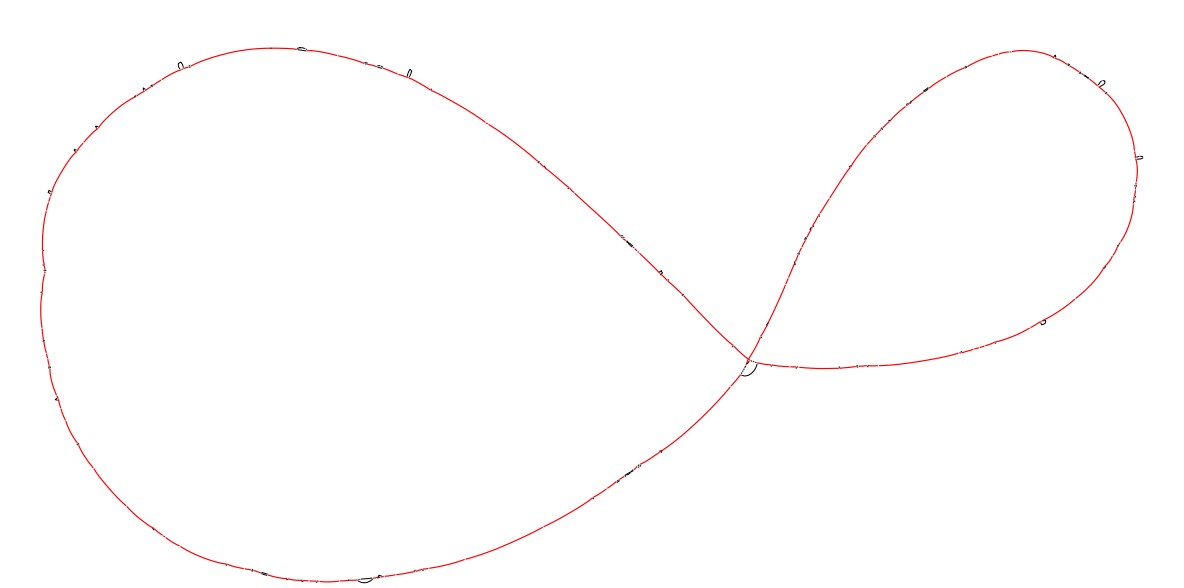
As expected the marginalized pangraph contains fewer blocks than the original one (388 vs 1244), and blocks are on average longer (mean length: 14 kbp vs 6 kbp). Blocks that appear in red are shared by both strains, while black blocks are present in only one of the two strains. The pangraph is composed of two stretches of syntenic blocks, which are in contact in a central point. This structure can be understood by comparing the two chromosomes with a dotplot. Using D-Genies on the two sequences[2] we obtain the following:
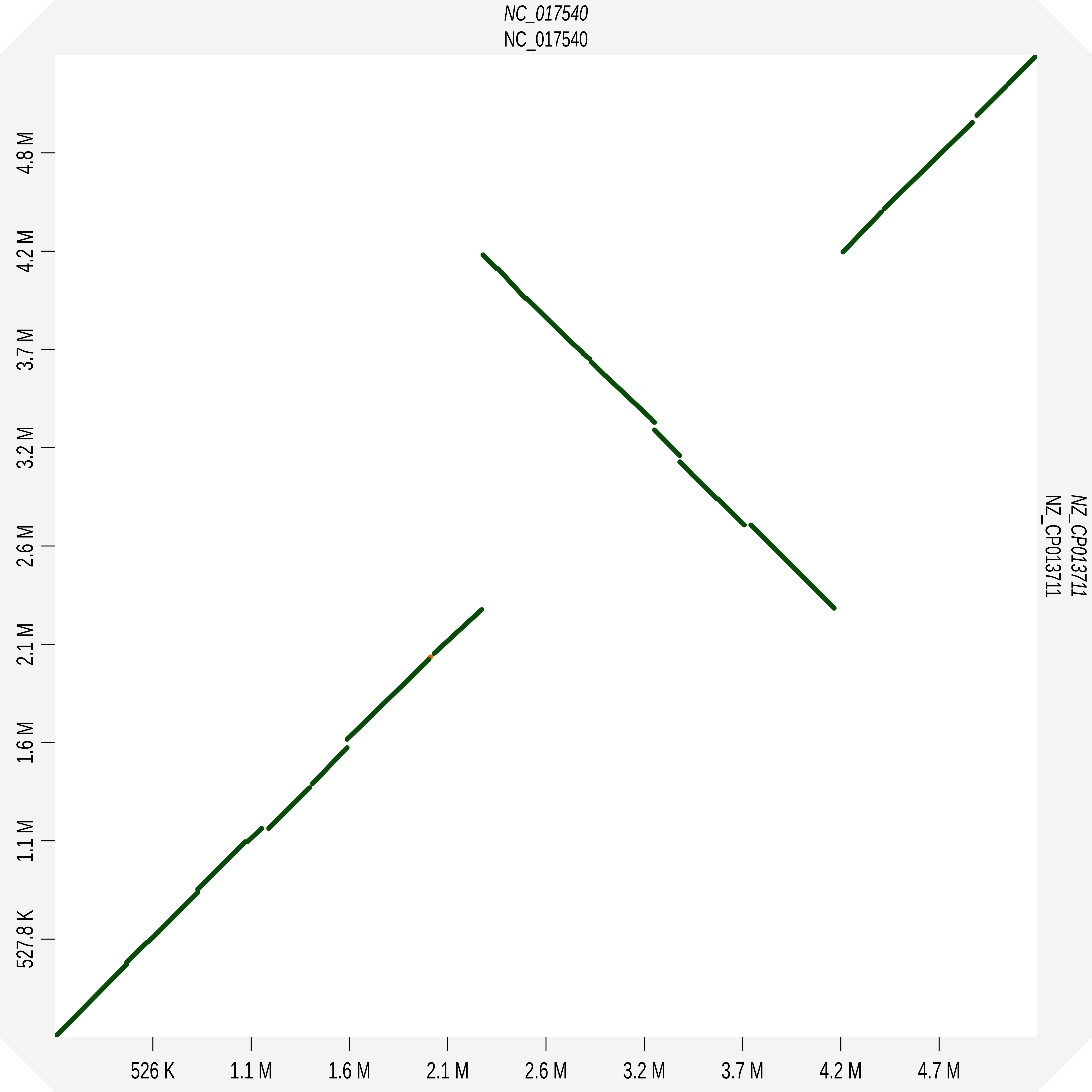
The contact point between the two loops in the pangraph is caused by the fact that the two genomes are composed of two mostly syntenic subsequences (the two loops) but these loops are concatenated with two different strandedness in the two strains. If we were to draw the two paths (relative to the two chromosomes) with different colors on top of the pangraph we would observe something similar to this: